The Wollman lab research spans development of new measurement technologies, creation of theory to understand the principles of organ architecture, and with many collaborators, apply these tools to understand disease
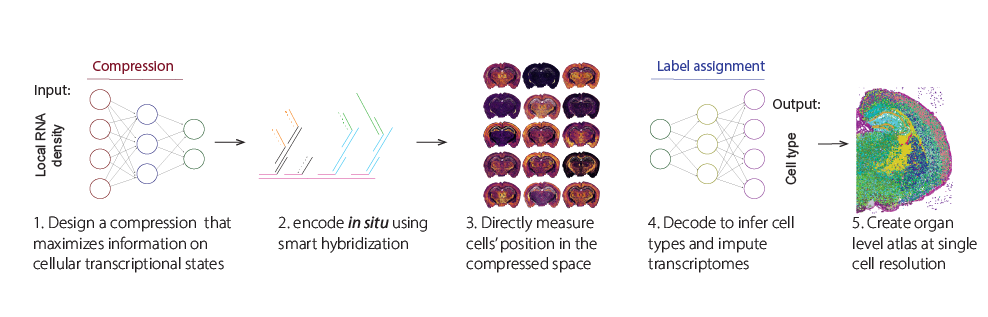
Using ATLAS we mapped the mouse brains of 15 animals with over 400 full coronal sectios infering cell types and imputing trascriptomes of over 40,000,000 cell.
With this many cells, we can reconstruct cell type specific spatial distribution of hundreds of cell types across different genetic backgrounds
- Theory: How do you build an organ with Lego-like bricks (cells) that differ from one another? How different are they—due to gene expression "noise" or functional heterogeneity? Is tissue architecture, the spatial arrangement of different cell types, inherently "noisy," and does this variability affect tissue function? What are the most informative ways to present organ atlases, and how does this change our understanding of organ organization? We tackle these questions using information theory and mathematical modeling to decode the relationships between cellular diversity, spatial organization, and organ functionality.
𝑆𝑅𝐼(𝐺𝑖,𝐺𝑗,𝐶𝑎2+)=𝐼(𝐺𝑖,𝐺𝑗,𝐶𝑎2+)−𝐼(𝐺𝑖,𝐶𝑎2+)−𝐼(𝐺𝑗,𝐶𝑎2+)